■ 基本信息
启动子: | 2×35s |
复制子: | pUC |
原核抗性: | Kan |
真核抗性: | Hyg |
克隆菌株: | DH5a |
培养条件: | 37度 |
■ 质粒属性
质粒宿主: | 植物 |
质粒用途: | 基因编辑 |
片段类型: |
|
片段物种: |
|
原核抗性: | Kan |
真核抗性: | Hyg
|
荧光标记: |
|
■ 质粒简介
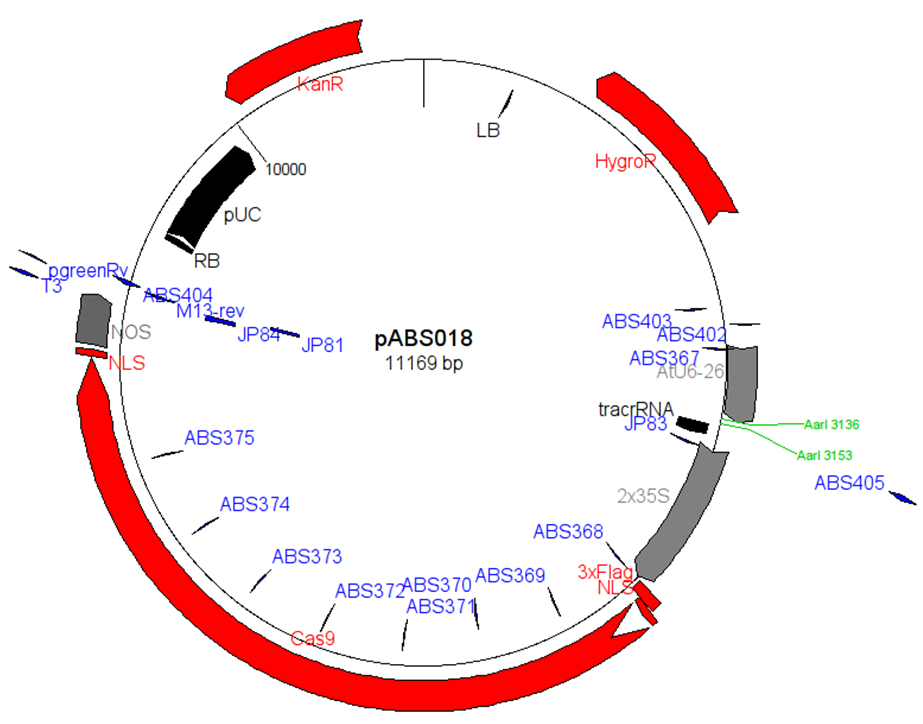
pABS018是一个植物细胞CAS9和gRNA表达质粒。This cloning procedure is very rapid, since the AarI enzyme cuts outside of its recognition site, the overhangs are incompatible and the subsequently-cloned in fragment does not restore the AarI site. Therefore, no purification is required to remove the restriction enzyme prior to ligation. In my experience, more than half the clones contain the desired insert.
Based on: https://groups.google.com/forum/#!msg/crispr/ofvc1KLCMfo/pf7jbjZP9FUJ
Cost, G.J., and Cozzarelli, N.R. (2007). Directed assembly of DNA molecules via simultaneous ligation and digestion. BioTechniques 42, 84, 86–89.
A. design oligos for cloning
Requirements for a good sgRNA: needs to be 20bp long, followed by NGG. The 20 bp sequence should have as many mismatches as possible with other genes to reduce off-target effects. It should also start with a G (this G may be required for U6 promoter function):
5'-GATTGNNNNNNNNNNNNNNNNNNN3'- (oligo #1)
||||||||||||||||||||
3'-CNNNNNNNNNNNNNNNNNNNCAAA-5' (oligo #2)
No need to order oligos phosphorylated so long as you don't de-phosphorylate vector.
*This website is excellent for helping choose a good target site:
http://www.genome.arizona.edu/crispr/CRISPRsearch.html
B. Anneal oligos to make double-stranded insert with sticky ends:
1. Mix both oligos together in 1x PCR buffer to 10 μM concentration
20 μL oligo1 (100 μM)
20 μL oligo2 (100 μM)
20 μL 10x PCR buffer
140 μL H2O
2. Float tubes in a beaker of 100-200 ml boiling water. Allow to cool several hours to room temperature. This ensures that annealing is complete.
C. AarI digest pABS018 plasmid (Concurrently with step B)
x μL pABS018 (1μg)
2 μL AarI RE buffer (10x)
1 μL AarI enzyme (2units/μL)
y μL H2O (to 20μL final volume)
→37°C for 1.5 hours
D. Ligate insert + plasmid
To 20μl digested plasmid, add:
2.5 μL 10x T4 Ligase Buffer
1 μL annealed oligo
1.5 μL T4 DNA Ligase
→37°C for 1.5 hours
E. Transform
Transform 2μL of ligation. Plate on Kanamycin Plates
F. Screen colonies and sequence
Doing colony PCR gives many false positives due to contamination by the ligation reaction. We get around this problem by replica plating the colonies onto new plates, and then doing colony PCR the following day. Alternatively you can PCR flaking the insert and then AarI digest. Those with the insert are AarI-resistant, but background colonies containing the original AarI site are cut by AarI.
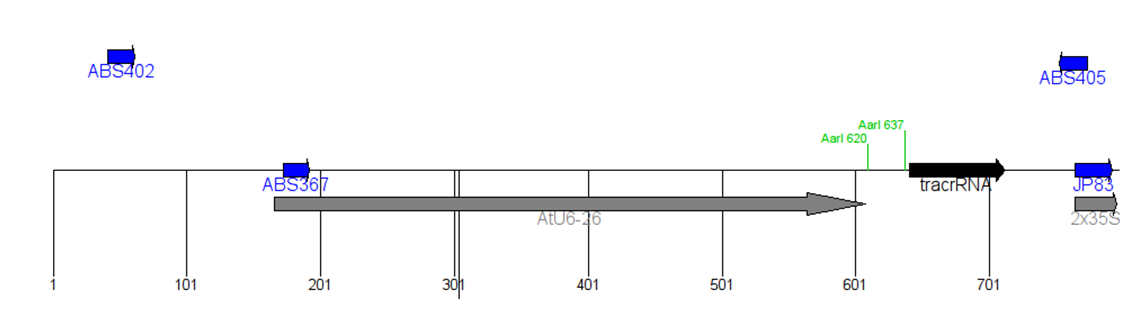
ABS 367 GTTGAACAACGGAAACTCGA
ABS 402 GTTTTCCCAGTCACGACGTTG
ABS 405 ATGCAAGCTTGCGGCCGCGCTG
For colony PCR to check for insert (on replica-plated colonies), use ABS367 and your sgRNA reverse primer. This will give a PCR product of ~468 bp.
For colony PCR to flank the insert, use ABS402 and ABS 405. This will give a PCR product of 732 bp. Then sequence that product with ABS 367.
■ 质粒图谱